Role of MiRNAs in Oral Cancer-Juniper publishers
JUNIPER PUBLISHERS-OPEN ACCESS JOURNAL OF HEAD NECK & SPINE SURGERY
Abstract
Oral cancer, one of the most common cancers
worldwide, exhibits high mortality and morbidity rates. The incidence
rate of oral cancer is high in developing countries, especially in Asian
countries. MiRNAs are non-coding RNAs with significant regulatory
functions, such as mRNA degradation or translation inhibition.
Understanding of the tumorigenesis of oral cancer has significantly
progressed at multiple levels. MiRNAs play an important role in oral
tumors and have been associated with specific oral cancer phenotypes,
such as tumorigenesis, progression, recurrence, or postoperative
survival. MiRNAs exist stably in common body fluids and are thus ideal
biomarkers for oral cancer. The miRNA profiles hallmark a potential
diagnostic value for detection of oral cancer and potentially malignant
disorders. In this review, we will summarize our current knowledge
regarding the most active miRNAs involved in oral cancer, focusing our
discussion on their role in tumor diagnosis, development, and
progression.
Keywords:MiRNAs; Oral cancer; Tumorigenesis; Biomarker; Diagnosis; Prognosis
Oral cancer, also known as “oral cavity cancer,” is
the sixth most common cancer worldwide and has high mortality and
morbidity rates. Oral squamous cell carcinoma is the most common type,
accounting for approximately 90% of oral cancers [1]. The 5-year
survival rate of oral cancer is approximately 50%, and more than 11,000
deaths are recorded annually [2]. Approximately 500,000 individuals are
diagnosed annually worldwide, and the incidence rate continuously
increases [3]. Oral cancer is prevalent in developing countries,
especially in South Asia [2]. Oral cancer is significantly caused by
environmental carcinogens, particularly smoking, alcohol, and betel quid
chewing [4]. Patients with oral cancer experience poor quality of life
because of the side effects of clinical treatments and cosmetic
problems. Oral cancer is a multi-step disease, but its precise mechanism
remains unknown [5]. Therefore, the pathogenesis of oral cancer and its
earlier biomarkers must be further explored.
MiRNAs are non-coding RNAs with approximately 17 to
25 nucleotides in length [6]. The biogenesis of miRNAs begins with
transcription of pri-miRNAs by RNA polymerase II. Pri-miRNAs are then
cleaved by Drosha in the nucleus, producing pre-miRNAs [6,7]. Pre-miRNAs
are subsequently exported into the cytoplasm and processed by Dicer to
form mature ∼21bp miRNAs, which are incorporated into RNA-induced
silencing complex [6,8]. Mature miRNAs bind to complementary sequences
of conserved
3′untranslated regions (UTRs), repressing translation and promoting
degradation of target genes (Figure 1) [6,9-11].

In recent decades, miRNAs have been regarded as
important factors for tumorigenesis [5,12]. Evidence suggests that
miRNAs function as either tumor suppressors or oncogenes [13,14].MiRNAs
are differentially expressed in distinct tumors and
involved in important tumor-related cellular programs, such
as cell proliferation, cell survival, cell differentiation,
antiangiogenesis,
invasion, or metastasis [15].
Many studies reported that miRNAs are associated with
specific oral cancer phenotypes, such as tumorigenesis,
progression, recurrence, or postoperative survival [5,16]. In
this review, we summarize commonly known miRNAs related
to oral cancer and discuss their biological functions or cellular
mechanisms [5] (Table 1). In addition, the stable existence of
miRNAs in common body fluids makes them ideal biomarkers
for diagnosis or prognosis of oral cancer.

Let-7b
Let-7b miRNA, one of the first discovered miRNAs, is a key
regulator for cell proliferation and differentiation [17,18]. Let-
7b miRNA is found in many cancers, including colon, pancreatic
endocrine, acinar tumors, and pancreatic adenocarcinoma
[19]. Andrew et al. [20] found that let-7b, but not let-7a, was
significantly reduced in oral cancer cell lines compared with
control cells. Over-expression of let-7b significantly reduced
the expression of its target gene Dicer, which is an RNase III
endonuclease required for miRNA maturation and aberrantly
expressed in different types of cancer [20]. Another study found
that let-7b also targets insulin-like growth factor 1 receptor
(IGF1R), which is activated by IGF1 or IGF2 through autocrine
and paracrine signaling. Let-7b inhibits cell proliferation
and colony formation and triggers S/G2 cell cycle arrest by
targeting IGF1R and IRS-2 through the Akt pathway [1]. Hence,
let-7b down-regulation contributes to oral cancer progression.
Over-expressing let-7b could be a potential therapy target for
inhibition of cancer cell proliferation.
miR-7/miR-21
MiR-7 and miR-21 are two keratinization-associated miRNAs
that are up-regulated in keratinized tumors compared with
normal or non-keratinized tumors [21]. Jung et al. [22] found
that miR-7 and miR-21 are up-regulated; in silico analysis results
indicated that RECK is the optimal candidate target gene for miR-
7 and miR-21. RECK plays crucial roles for tumor progression
by degrading the extracellular matrix barriers, encompassing
the tumor and permitting invasion into surrounding connective
tissues. In oral cancer cells, miR-7 and miR-21 are inversely
correlated with the expression of RECK [22]. Patricia et al. [23]
also found that miR-21 is up-regulated in oral cancer; this study
revealed that miR-21 directly binds to the 3′UTR of PDCD4,
which is associated with disease progression and metastasis
[23]. Further study showed that the expression of miR-21 is the
major factor related to the poor prognosis of patients with oral
cancer [24]. Overall, miR-7/miR-21-induced deregulation of
RECK may contribute to the aggressiveness of tumors.
miR-99a
MiR-99a is characterized as tumor suppressor in several
human cancers, including childhood adrenocortical tumors,
prostate cancer, liver cancer, head and neck cancer, and oral
cancer [25-27]. MiR-99a is one of the most down-regulated
miRNAs in oral cancer cell lines compared with normal oral
keratinocytes. Transwell and tail vein injection assays indicated
that miR-99a functions in tumor migration/invasion and lung
colonization. Moreover, target prediction and real-time PCR
assay validated that IGF1R is the target gene of miR-99a, which is
a transmembrane tyrosine kinase receptor [28]. The expression
of miR-99a is negatively correlated with the expression of IGF1R
in oral cancer cells. Activation of IGF1R leads to activation of
the Ras, Raf, and mitogen-activated protein kinase pathway
and thus enhances the proliferation and stimulation of the
phosphatidylinositol 3-kinase pathway, resulting in apoptosis
inhibition [29]. Collectively, miR-99a mutually regulates its own
target, IGF1R, suggesting the possibility of miR-99a for targeting
IGF1R in cancer therapy.
miR-100
MiR-100 is down-regulated in many cancers, including
ovarian cancer and hepatocellular carcinomas [30,31]. Genetic
aberrations are common in oral cancer, and chromosome 11q
is among the most common alterations [32,33]. Brian et al.
[34] examined the expression of miRNAs mapped to 11q in oral
cancer cell lines compared with normal human oral keratinocytes
(NHOK); results indicated that mir-100 is down-regulated in
oral cancer cells. Cell proliferation is significantly reduced when
exogenous miR-100 is over-expressed. Microarray analysis data
revealed that over-expression of exogenous miR-100 downregulated
a number of target genes including ID1, EGR2, MMP13,
and FGFR3; these genes are involved in cell metastasis, myelin
development, cell adhesion, and cell growth, respectively. In
conclusion, miR-100 inhibits cell proliferation by targeting
several key genes involved in cancer and plays an important role
in the development and progression of oral cancer [34].
miR-125
MiR-125b is also located in chromosome 11q and involved in
important cellular processes, especially in neuronal development
and differentiation [33]. The alteration of miR-125b occurs
in many cancers, including ovarian cancer, breast cancer, and
prostate cancer [35,36]. Consistent with miR-100, miR-125b is
also down-regulated in oral cancer cell lines. Over-expression
of exogenous miR-125b significantly reduces cell proliferation.
In oral cancer, miR-125b regulates the expression of key factors
involved in tumorigenesis; these factors include KLF13, CXCL11,
and FOXA1. All these target genes have important chemokines
and transcription factors, which have crucial biological
functions in cell growth, cell proliferation and differentiation,
inflammation, angiogenesis, and metastasis [31,34]. Shiiba
et al. [16] also found that miR-125b is down-regulated in oral
cancer cell lines, resulting in increased cell proliferation rate
and decreased radio-sensitivity to X-ray irradiation; this study
reported a new target of miR-125b, namely, ICAM2, which is
involved in cell proliferation [16]. All these lines of evidence
suggest that miR-125b contributes to the development and
progression of oral cancer by regulating important target genes.
Thus, understanding of miR-125b increases our knowledge of
the molecular mechanisms underlying oral cancer. Furthermore,
miR-125b may be a potential therapeutic target for oral cancer.
miR-145
MiR-145 acts a tumor suppressor in numerous human
cancers, including prostate cancer, bladder cancer, and colon
cancer [37-39]. A study found that miR-145 is down-regulated
in oral cancer tissues compared with normal mucosa tissues.
Exogenous over-expression of miR-145 induced G1 phase arrest
and cell apoptosis, which then suppressed cell proliferation and
colony formation. The two direct targets of miR-145 are c-Myc
and Cdk6, which are important oncogenes. In oral cancer cells,
miR-145 also targets c-Myc and Cdk6, leading to inhibition of oral
cancer cell growth [40]. Thus, miR-145 rescue may be a rationale
for diagnostic and therapeutic applications in oral cancer.
miR-146a
MiR-146a is suppressed in many malignancies [41] but has
been recently identified to be up-regulated in oral cancer [42].
Hung et al. [43] found that miR-146a exhibits higher expression
in oral cancer than matched adjacent mucosal cells. Inhibition
of miR-146a significantly blocks the growth of xenograft tumors.
miR-146a directly targets IRAK1, TRAF6, and NUMB, thereby
enhancing cell proliferation, invasion, and metastasis. The plasma
miR-146a levels of patients with oral cancer are significantly
higher than those of control subjects; hence, miR-146a could be
a potential biomarker for diagnosis of oral cancer [43].
miR-155
MiR-155 is overexpressed in many human cancers, including
lymphoma and solid tumors of diverse origin (e.g., breast, lung,
stomach, prostrate, colon, thyroid, and pancreatic) [14,44,45].
MiR-155 is also up-regulated in oral cancer [46]. In oral
cancer, miR-155 targets CDC73, which negatively regulates
β-catenin, cyclin D1, and c-MYC. Ectopic expression of miR-155
dramatically reduced CDC73 levels, increased cell viability, and
decreased apoptosis in nude mice. Conversely, inhibiting miR-
155 resulted in increased CDC73 levels, decreased cell viability,
and increased apoptosis. Hence, the tumor suppressor CDC73 is
a target of oncogenic miR-155 [47]. Furthermore, the reversal
of pro-oncogenic properties of miR-155 due to its inhibitor is a
promising method for cancer therapeutics [47].
miR-196
MiR-196 is an active molecule and is altered in several
cancers. Studies stated that miR-196 is down-regulated in
melanoma and acute lymphoblastic leukemia [48]. However,
other scholars reported that miR-196 is up-regulated in several
malignant diseases, including esophageal cancer, pancreatic
cancer, colorectal cancer, and glioblastoma [49]. Lu et al. [50]
examined the expression of miR-196 in oral cancer cell lines and
normal keratinocyte cell lines; results indicated that miR-196
is highly up-regulated in oral cancer cells. In vitro experiments
showed that miR-196 is responsible for cell growth, migration,
invasion, and radio/chemosensitivity. Target prediction and
subsequent validation experiments identified that miR-196
targets NME4, a member of the NM23 family. NME4 then regulates
the downstream JNK-TIMP1-MMP signaling pathway, which
plays an important role in cell mobility [50]. In conclusion, miR-
196 promotes invasive and migratory phenotypes in oral cancer
by targeting NME4, leading to the regulation of the downstream
JNK-TIMP1-MMP signaling pathway [51].
miR-205
Various studies examined the biological functions
of microRNA-205 (miR-205) as a tumor suppressor in
carcinogenesis [52]. Kim et al. [53] found that miR-205 is significantly down-regulated in human oral cancer compared
with NHOKs. Reduced miR-205 expression in oral cancer cells
might be important for oral cancer progression. Over-expressing
exogenous miR-205 reduced cell viability in oral cancer cells and
induced cell apoptosis by activating caspase-3/-7. In addition to
caspase-3/-7, over-expressed miR-205 significantly promotes
the expression of IL-24, a well-known cytokine that functions
in cell apoptosis. IL-24 promoter scanning analysis revealed a
potential binding sequence located at position-151 in the IL-24
promoter. miR-205 interacts directly with the promoter target
sequence of IL-24 [53]. Some studies stated that miRNA can upregulate
the expression of their own target genes through an
interaction with the regulatory region [54,55]. These findings
indicate that miR-205 functions as a tumor suppressor in upregulating
IL-24 by targeting specific sites in its promoter. Thus,
miR-205 exhibits a significant therapeutic potential as molecular
medicine for treatment of oral cancer.
miR-483-3p
MiR-483-3p is involved in the final stage of skin wound
healing and arrest of keratinocyte proliferation [56]. MiR-483-3p
is down-regulated in oral cancer cells. Over-expressing miR-483-
3p significantly hinders tumor growth. Target prediction analysis
and validation experiments revealed that miR-483-3p targets
API5, RAN, and BIRC5, which play a main role in cell apoptosis.
MiR-483-3p exhibits its pro-apoptotic activity by binding to
these target genes, leading to inhibition of cell proliferation
and increase in cell apoptosis [57]. Hence, miR-483-3p may be
used as an adjuvant in many cancers characterized by downregulation
of miR-483-3p.
miR-518c-5p
MiR-518c-5p belongs to the miR-515 family and is located
in chromosome 19. MiR-518c-5p is originally identified in 26
different organ systems of humans and rodents and is enriched
in neuronal as well as normal and malignant hematopoietic
cells and tissues [58]. However, the function of miR-518c-5p in
cancers remains unclear. Makoto et al. [59] found that SDF-1/
CXCR4 system is involved in the establishment of metastasis in
oral cancer. Subsequent examination showed that miR-518c-
5p is induced and exhibits distant metastatic potential. When
inhibiting the function of miR-518c-5p by using LNA-modified
inhibitors, the cell growth and migration of oral cells are reduced
significantly. Consistently, over-expression of miR-518c-5p
enhances the migration of oral cancers. An in vivo experiment
indicated that exogenous miR-518c-5p significantly increased
the tumor volume, lymph node metastasis, and lung metastasis
in mice. The miR-518c-5p inhibitor may serve as an ideal target
for development of therapies against metastasis [59]. However,
the target genes of miR-518c-5p remain unknown. Future
studies must critically examine the target genes of miR-518c-5p
in oral cancer.
Early detection of potentially malignant oral cancer is
important for improving the probability of complete recovery
because the stage of malignancy at the time of diagnosis
influences morbidity and mortality [60]. However, more than
60% of patients present with stage III and IV oral cancer. As such,
scholars must explore the most potential biomarkers for early
detection of oral cancer (Table 2) [60].
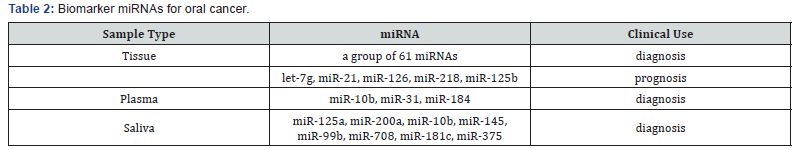
Lajer et al. [61] compared 51 patients with oral cancer with
40 control patients by using microarray analysis and found
114 differentially expressed miRNAs. A molecular classifier
including 61 miRNAs was generated for diagnosis of oral cancer;
this classifier exhibits 93% accuracy, 100% sensitivity, and 86%
specificity [61].
Peng et al. [62] examined the profile of miRNA from
58 oral cancer samples and paired normal tissues through
microarray assay. A total of 232 of the 760 miRNAs assessed
were differentially expressed between paired tumor and normal
tissue samples. Among the 232 miRNAs, the reduced expression
of miR-218, miR-125b, and let-7g is associated with high risk of
poor outcomes [62]. MiR-21 is a common biomarker for many
cancers. A previous study on 86 patients with oral cancer revealed
that miR-21 was primarily expressed in the tumor stroma but
was not expressed in the adjacent normal epithelia. Multivariate
Cox-regression analysis of disease-free survival revealed that
miR-21 expression level was significantly associated with poor
prognosis [63]. Sasahira et al. [64] found that decreased miR-126
expression was strongly correlated with disease-free survival
in 118 cases with oral cancer. Another multivariate analysis of
patients with oral cancer indicated that a poor prognosis was
associated with low miR-126 expression compared with high
miR-126 expression (P=0.0013) [64].
Some biological molecules are present in body fluids, such
as plasma and saliva, and are ideal noninvasive biomarkers
for oral cancer [65]. Previous studies used protein, mRNA, and
DNA extracted from body fluids to detect oral cancer. Analysis
using transcriptomic and proteomic technology discovered and validated mRNA and protein salivary biomarkers, respectively, to
be highly discriminatory for oral cancer detection. Plasmic and
salivary miRNAs are protected from ribonucleases present in the
plasma/saliva by macromolecules called exosomes, which are
known to package and transport miRNAs [65-67]. The presence
of miRNAs in human body fluids, especially in saliva, is an
emerging field for monitoring oral diseases.
Liu et al. [68] identified that miR-31 in plasma was
significantly elevated in patients with oral cancer (n = 43)
compared with control individuals (n = 21). Plasma miR-31
yielded a receiver operating characteristic curve area of 0.82 and
an accuracy of 0.72 defined by leave-one-out cross-validation
[68].
Wong et al. [69] profiled the expression of 156 miRNAs
on four oral cancer and paired normal tissues, revealing 37
differentially expressed miRNAs (three-fold). Among the 37
miRNAs, miR-184 was further examined in the plasma from 20
patients with oral cancer and normal individuals. Plasma miR-
184 level was significantly higher in patients with oral cancer
and s, and significantly decreased after surgical removal of the
primary tumors [69]. Lu et al. [70] found that miR-10b was
considerably elevated in the plasma of patients with oral cancer
[P < 0.0001, area under curve (AUC) = 0.932] and pre-cancer
lesions (P < 0.0001, AUC = 0.967); hence, miR-10b could be a
potential early diagnostic biomarker for oral cancer [70].
Park et al. [66] examined and compared the expression of
approximately 50 miRNAs in whole saliva and saliva supernatant
between patients with oral cancer and healthy controls. Two
saliva miRNAs, namely, miR-125a and miR-200a, were present in
significantly lower levels (P<0.05) in the patients than that in the
controls [66]. Yang et al. [71] first examined the profile of saliva
miRNAs in 10 patients who developed carcinoma and 35 patients
who did not develop oral cancer; results indicated that miR-10b,
miR-145, miR-99b, miR-708 and miR-181c were significantly
differentially expressed [71]. Wiklund et al. [72] used TaqMan
RT-PCR arrays to profile miRNA expression in saliva from 15
patients with oral cancer and 7 healthy controls; the results
showed that miR-375 was down-regulated in the patients and
thus could be used to diagnose oral cancer [72].
Oral cancer is the sixth most common cancer worldwide
and has high mortality and morbidity rates. Understanding the
tumorigenesis and development of oral cancer has significantly
progressed. The expression of many miRNAs is altered in oral
cancer, thereby regulating cell growth, cell proliferation and
apoptosis, invasion, and metastasis. Accumulating evidence
highlights that circulating miRNAs are significantly associated
with oral cancer and thus could be used as potential biomarkers
for diagnosis and prognosis of oral cancer [73-75]. However, the
highly stability makes miRNA ideal biomarkers for non-invasion
detection. Further work is required to elucidate the complete
regulatory mechanisms of miRNAs. The clinical use of miRNA as
biomarkers needs more tests and more patients to validate the
most potential markers.
To know more about Open Access Journal of
Head Neck & Spine Surgery please click on:
To know more about juniper publishers: https://juniperpublishers.business.site/
Comments
Post a Comment